Info
If you use this website in a publication, please cite the CHASMplus paper:
Tokheim and Karchin, CHASMplus Reveals the Scope of Somatic Missense Mutations Driving Human Cancers, Cell Systems (2019), https://doi.org/10.1016/j.cels.2019.05.005
Summary
Large-scale cancer sequencing studies of patient cohorts have statistically implicated many cancer driver genes, with a long-tail of infrequently mutated genes. Here we present CHASMplus, a computational method to predict driver missense mutations, which is uniquely powered to identify rare driver mutations within the long-tail. We show that it substantially outperforms comparable methods across a wide variety of benchmark sets. Applied to 8,657 samples across 32 cancer types, CHASMplus identifies over 4,000 unique driver mutations in 240 genes, further distinguished by their specific cancer types. Our results support a prominent emerging role for rare driver mutations, with substantial variability in the frequency spectrum of drivers across cancer types.
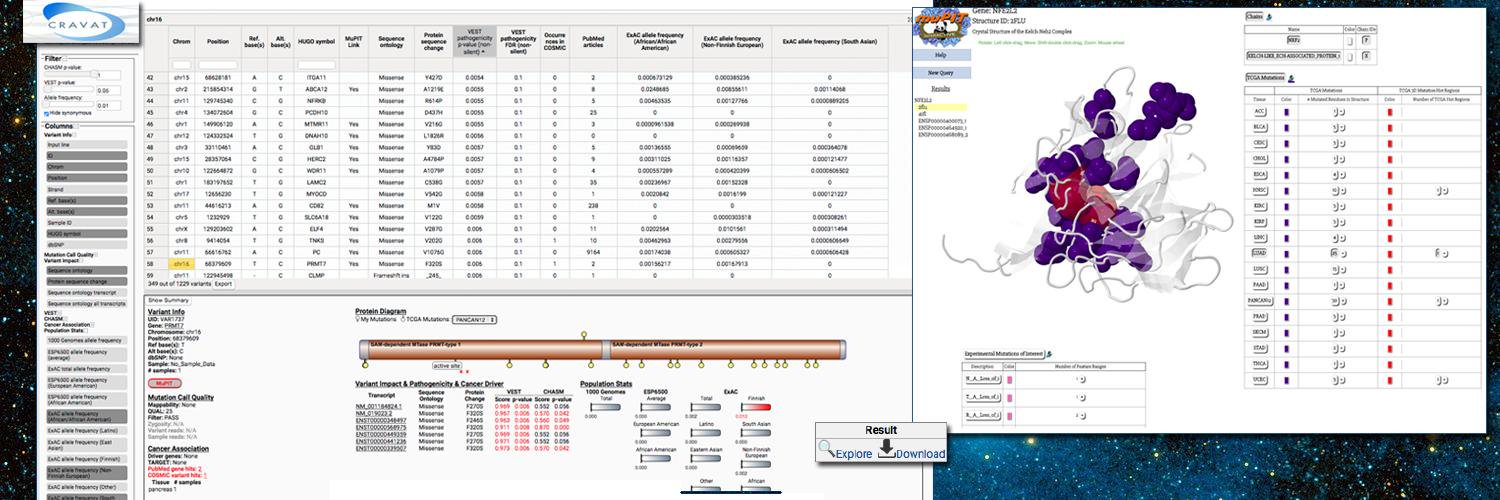
Given the sheer number of driver mutations identified by CHASMplus, we created an interactive resource so that non-bioinformaticians could further explore our results. Results are viewable through the interactive results viewer by CRAVAT. This enables users to understand the predicted driver missense mutations in context with other mutations from The Cancer Genome Atlas and visualize results. As an example, mutations can be visualized at their corresponding position on 3D protein structures.
